miRNAtools3 > Tutorials > SNPs vs miRNAs
Scenario 4: SNPs and miRNA binding sites
![]()
1.- INTRODUCTION: working hypothesis - cannabinoid receptor
It is very well known that the genetic variants present in coding regions of different individuals are in the basis of several biological processes, including the propensity to disease or the therapeutic response to drugs. Moreover, the presence of genetic variants in the 3'-UTR region of genes is also a factor that can influentiate the genetic output, since it can be related with the effect of post-transcriptional regulators such as miRNAs. If a SNP is located in a miRNA binding site, a different regulatory effect from this miRNA between the genetic variants is expected. SNPs in miRNA binding sites have been studied in some biological context, and related for instance with the risk of breast cancer (See Zhang et al, 2011).
For this tutorial we will analyze an hypothetical case not yet biologically demonstrated, and related with the presence of SNPs in miRNA binding sites. In humans the CNR1 gene encodes for a membrane protein (cannabinoid receptor) which is on the basis of several biological processes like tha perception of pain, memory or apetite. This receptor is also the target of some chemical compounds present in plants that are used as recreative drugs. Let's hypothesize: are there any genetic variants that could be related with the miRNA regulation of CNR1 gene transcript, and consequently with the different tolerance to cannabinoid substances observed in different individuals?
2.- Searching for SNPs in the 3'-UTR of CNR1 gene
We will use miRdSNP (See the specialized databases section), a database which combines described SNPs in 3'-UTR regions of human genes together with predicted and validated miRNA targets, an excellent starting point for these kind of studies. For starting a search, open miRdSNP frontpage and go to the "Search" option in the menu above. You will get redirected to a webpage which contains all the available information in the database. In order to filter the results to search for the SNPs in the CNR1 gene, write "CNR1" in the "Gene" searching window. After that, your results will be filtered and presented in a table like that:
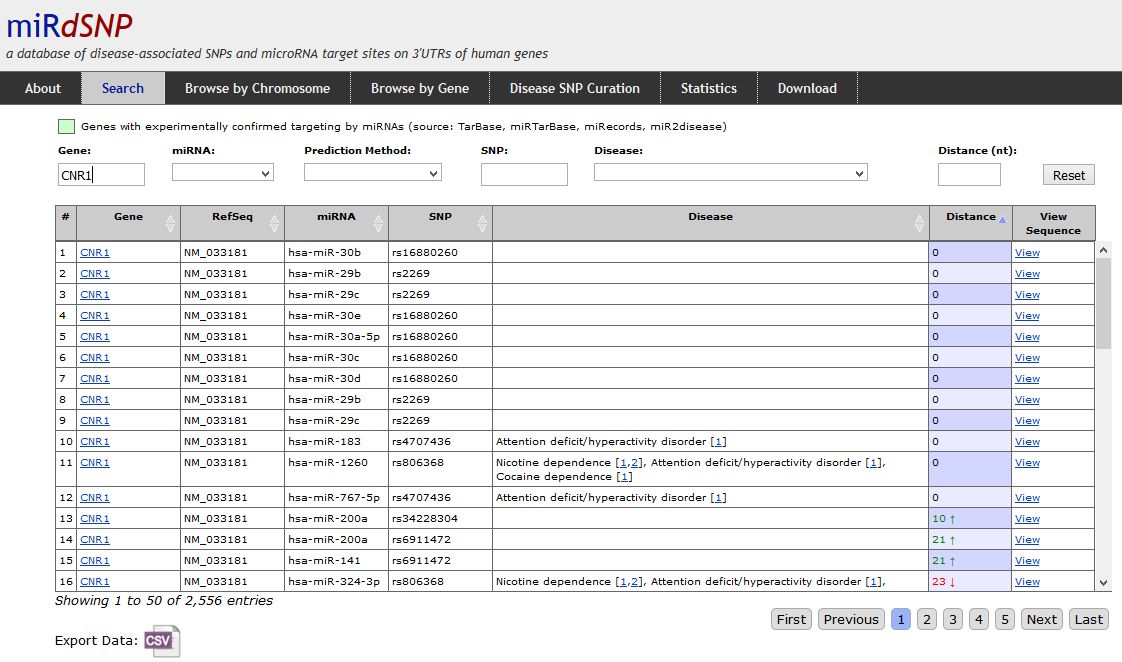 |
| Output results for the 3'-UTR SNPs in the CNR1 gene in miRdSNP. In the columns you will se information about the gene name, ref-seq entry, targetting miRNA, SNP reference and disease involved in this particular SNP. It is important to check the column named "Distance". This column displays the distance of the reported SNP to the corresponding miRNA target in nucleotides. Of course that if we want to look to SNPs that can have influence on miRNA binding, we should only look to the smaller numbers, tipically equal to zero. You can see that some of the SNPs have been already reported to be related with some diseases or conditions (Column "Disease"). |
3.- miRNA-mRNA hybrids: functional consequences of SNPs
The presence of SNPs in the binding regions for miRNAs can either result in a gain of a new target or a loss of a pre-existent regulatory sequence. This effect could have effects over the regulatory capacity of a particular miRNA. In fact, some of the already described SNPs in CNR1 gene have been related with human disorders or conditions by Genome Association Studies, but their functional consequences are in many cases unknown. In 2009, Zuo and coworkers published an article relating two CNR1 variants with increased risk for cocaine dependency (Zuo et al, 2009). One of this variants named rs806368 appeared in the table of results obtained with miRdSNP, and involves a C/T SNP in the 3´-UTR region of the CNR1 gene.
This SNP falls exactly in the target sequence for hsa-mir-1260a. In order to know what will be the effect of this SNP over the regulatory activity of this miRNA, we will take advantage of another database named miRNASNP2 (See the specialized databases section), which also compiles relationships between SNPs and miRNAs but is more updated than miRdSNP.
Open miRNASNP2 webpage and go to the "Search" option in the top menu. You will be redirected to a screen with many input options. In the bottom right section of the page you will see a square that will allow you query the database by SNPs in the UTR region of selected genes. Here, input the reference of the SNP (rs806368), and click in "Gain".
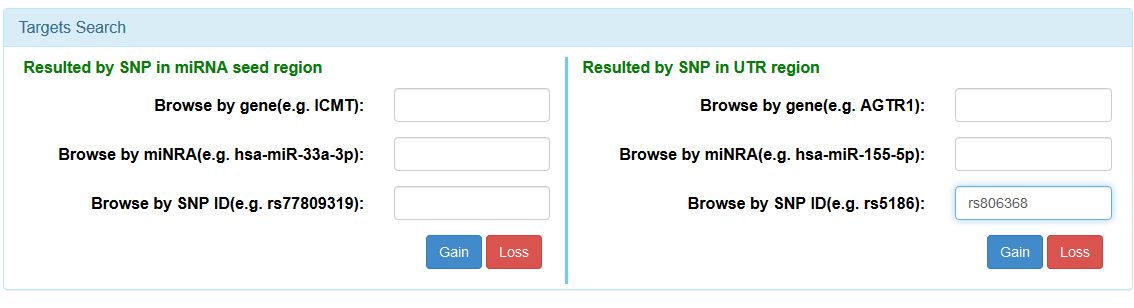 |
| Input section for targets search in miRNASNP2. To search for a specific SNP, input the reference as indicated and either click "Gain" or "Loss" if you want to chek if this SNP will have a positive or negative effect over the miRNA target recognition. |
After clicking "Gain" you will be redirected to a results page in a tabular format. In this table you will get the results about the influence of this SNP in the gain of function of miRNAs. You will see something like that:
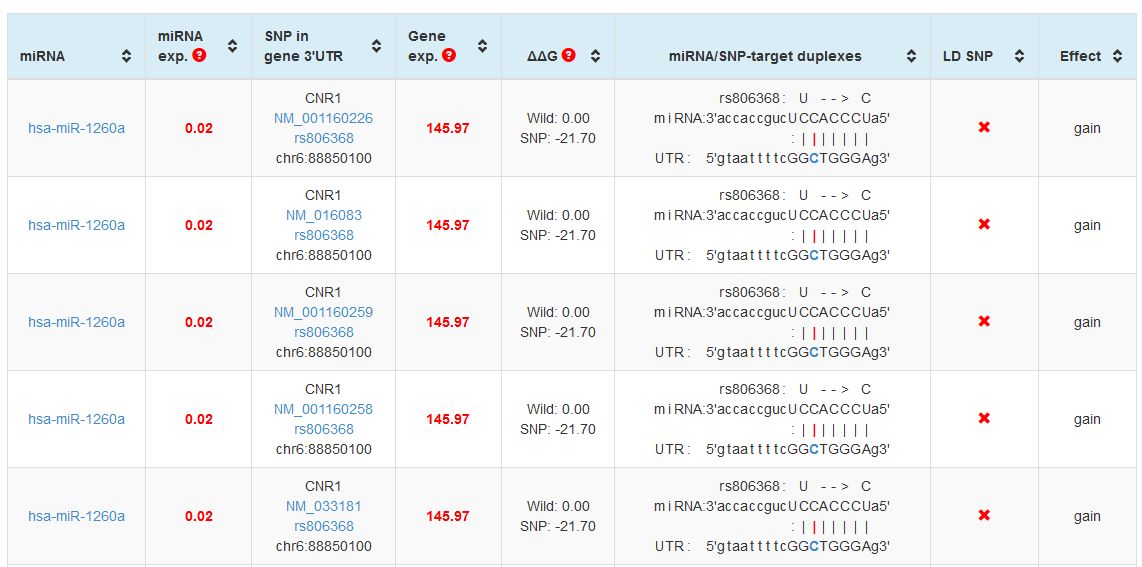 |
| Search output for the rs806368 SNP in miRNASNP2. The table shows the miRNAs which have gained function when this SNP appeared. As you can see by the left column, only one miRNA (hsa-mir-1260a) appeared. The repetition of the rows is due to the different isoforms of the CNR1 gene transcripts. You can also see in columns 2 and 4 the average expression values for the miRNA and mRNA in a cohort of tissues analyzed in the database. Interestingly you can see in the values of the DeltaG function of the miRNA-mRNA hybrids, that in the native situation there is no binding predicted for hsa-miR-1260a, whereas in rs806368 the free energy of the complex denotes a strong binding. It is a clear effect of a gain of function! |
Can you interpret the results?... What can you say about the action of CNR1 when rs806368 SNP is present?... What do you expect?...
Of course that for answering those interesting questions, you may need to take into account that the action of a particular miRNA is not possible if either the miRNA and the target are expressed in the same cell. So, be careful before taking crazy conclusions. You probably may check if CNR1 and hsa-miR-1260a are expressed in the cell that you are trying to study... You know how to do that. If not, check Scenario1.