miRNAtools3 > Tutorials > Single miRNA scenario
Scenario 1: Single miRNA
![]()
1.- INTRODUCTION: miR-21 as an oncomir
The work by Asangani et al in 2008, showed one of the first practical evidences of an oncomir (oncogenic miRNA) involved in cancer progression and metastasis. The authors were able to demonstrate the regulatory control of miR-21 exerter over the tumor supressor Pdcd4. A bioinformatics search revealed a conserved target-site for miR-21 within the Pdcd4-3'-UTR at 228-249 nt. In 10 colorectal cell lines, an inverse correlation of miR-21 and Pdcd4-protein was observed. Transfection of Colo206f-cells with miR-21 significantly suppressed a luciferase-reporter containing the Pdcd4-3'-UTR, whereas transfection of RKO with anti-miR-21 increased activity of this construct. This was abolished when a construct mutated at the miR-21/nt228-249 target site was used instead. Anti-miR-21-transfected cancer cells showed an increase of Pdcd4-protein and reduced invasion. Moreover, these cells showed reduced intra-vasation and lung metastasis in a chicken-embryo-metastasis assay. In contrast, overexpression of miR-21 in Colo206f significantly reduced Pdcd4-protein amounts and increased invasion, while Pdcd4-mRNA was unaltered. Resected normal/tumor tissues of 22 colorectal cancer patients demonstrated an inverse correlation between miR-21 and Pdcd4-protein. This is the first study to show that Pdcd4 is negatively regulated by miR-21. Furthermore, it was the first report to connect miR-21 with the induction of invasion, intravasation and metastasis.
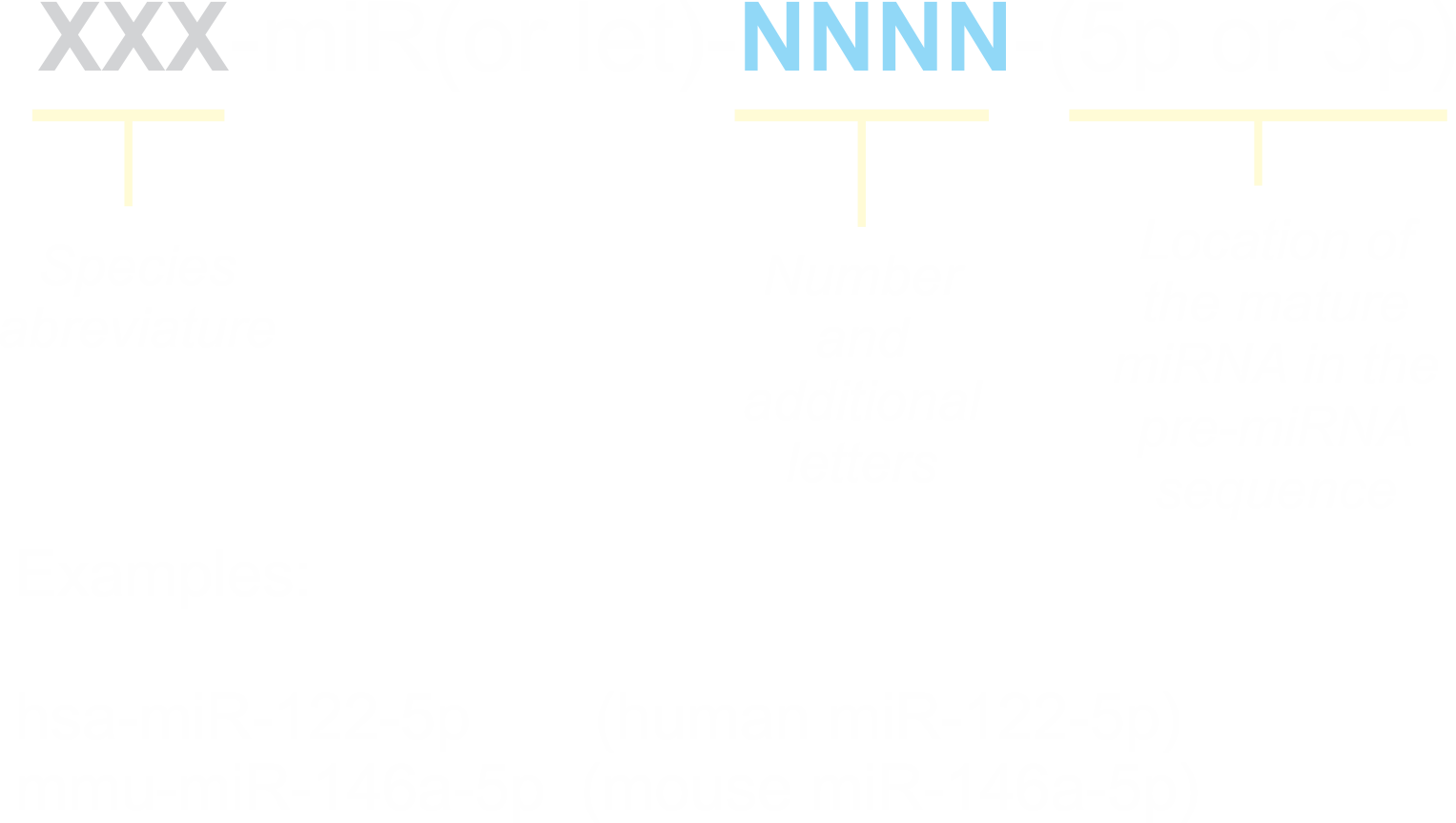
2.- Retrieving miR-21 data from miRBase: miRNA nomenclature
miRBase is the general database containing miRNA information. The actual version of miRBase (21.0) uses the following scheme for naming miRNAs:
 |
To retrieve information about miR-21 from miRBase (General Purpose Databases section), open its webpage and introduce the search keyword "hsa-mir-21" in the window for name or keyword search located in the right-hand side of the screen, and click "Go". You will get access to the information about the miRNA in a miRBase entry format (See miRBase entry for hsa-miR-21). Important fields in miRBase entries:
 |
Sequence of the precursor stem loop of hsa-miR-21, indicating both possible mature miRNAs generated from this loop. |
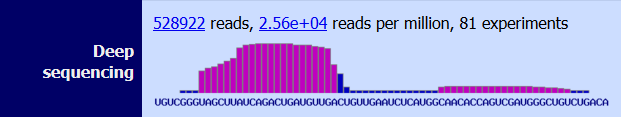 |
Abundance of each mature miRNA generated from the pre-miRNA as quantified by NGS in several experiments |
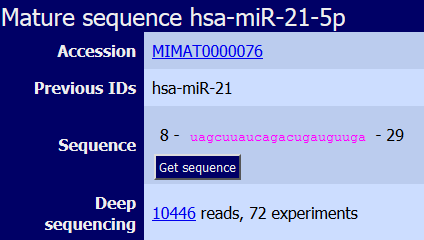 |
Sequence of the miRNA generated from the -5p end of the precursor. LOOK at the Previous ID to identify the old nomenclature of this miRNA. In fact, hsa-miR-21 corresponds to hsa-miR-21-5p in the last version of miRBase. |
 |
Sequence of the miRNA generated from the -3p end of the precursor. LOOK at the Previous ID to identify the old nomenclature of this miRNA. In old versions of miRBase the less abundant mature miRNA generated from the same precursor was designated as "STAR" (*). This nomenclature is currently OUT OF USE. |
To get information about the evolution of the nomenclature of a particular miRNA along the different miRBase versions, we can take advantage of the existence of applications such as miRBaseTracker or Miriadne (General Purpose Databases section). This is particularly useful when we are dealing with data published before 2010, where the miRNA nomenclature was not already standarized. Unfortunately, many journals do not take special care with this issue, and it is very frequent to see "old-fashion" miRNA names even in high-impact journals..... so, BE CAREFUL!
Here you can see the information for hsa-miR-21-5p in Miriadne under the "Time warp" search option:
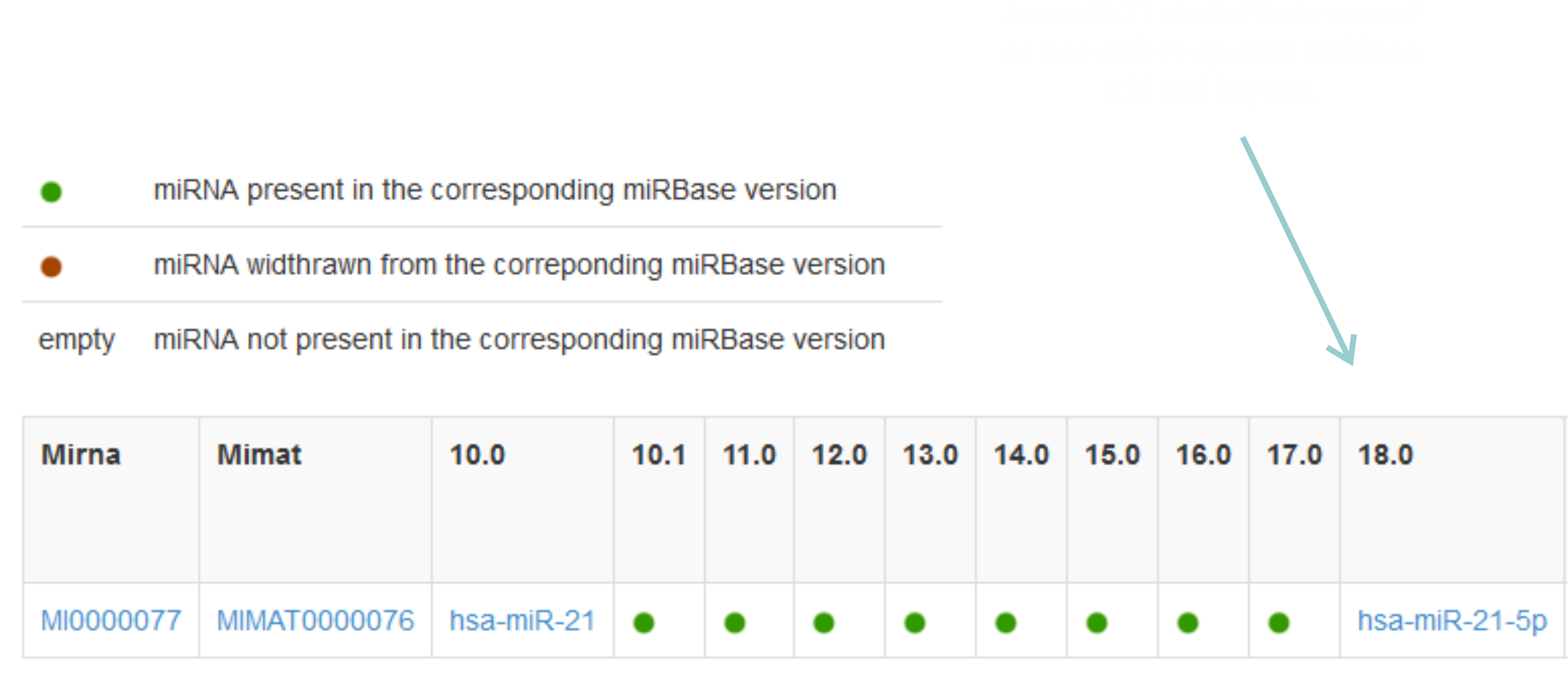 |
3.- Analyzing the expression profile of hsa-miR-21-5p in human tissues
In order to understand the physiology of miRNAs in several cellular contexts, it would be interesting to know its expression profile either in tissues or cells. For this objective we will use the miRmine application (General purpose databases section). Open miRmine and introduce the miRNA query "hsa-miR-21-5p" in the search window. Select "Tissues - All" to get the expression profile of this miRNA in a collection of human tissues. The results from miRmine are a compilation of data obtained from several NGS experiments. Here are the graphical representation of the search results in miRmine, but you can also get a table with numerical values easily importable into Excel:
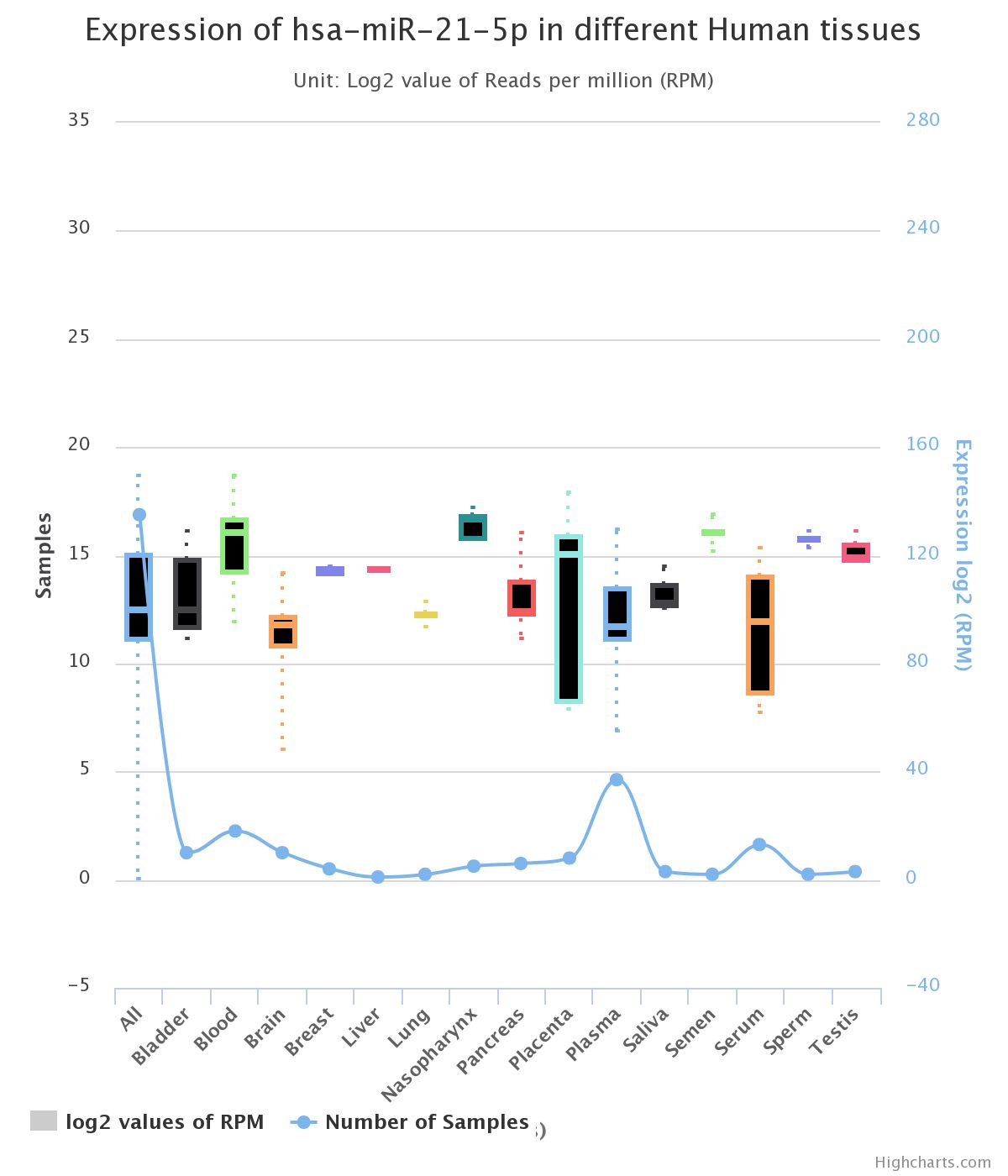 |
4.- Predicted targets for hsa-miR-21-5p
MiRNA target prediction is a complicated task, specially in animals, where the target is recognized by a short sequence composed by 7-8 nucleotides in the 5'-end of the miRNA (seed sequence). Many target prediction algorihms are available, and all of them have pros and cons. For this reason I strongly advice to use "multiple predictors", applications that simultaneously check many target prediction algorithms. For the purpose of this tutorial we will use mirWalk2 (See multiple predictors section). Open miRWalk2, open the "Predicted target module", and select "MicroRNA-gene targets". In the following window, fill the IMPORTANT searching options as indicated and push the SEARCH button:
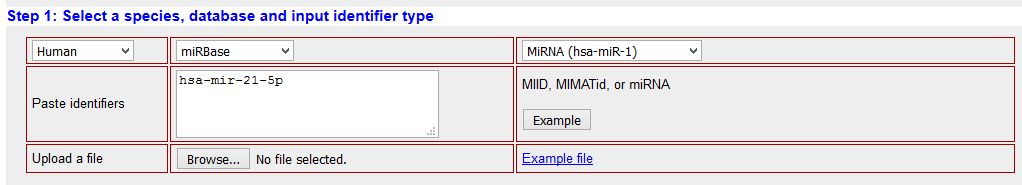 |
| STEP 1: selection of organism, database and nomenclature. BE CAREFUL in putting the right nomenclature for your miRNA according to the selection. If something goes wrong you will have to reset the search |
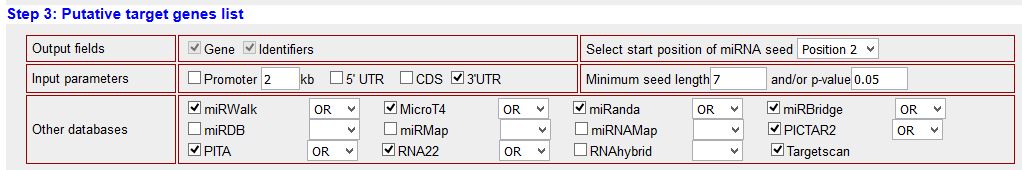 |
| STEP 3: selection of target prediction algorithms, searching sequence and seed parameters. We have selected to search only for the 3'-UTR, starting with the second nucleotide of the seed sequence (canonical binding) for a minimum seed lenght of 7 nucleotides. We have selected 8 target prediction algorithms. |
After the search you will get to a screen that will show you how to access the data related with the information about your miRNA, putative target tables and also something abut enriched functional patterns. Just let be focused in the putative target gene tables.
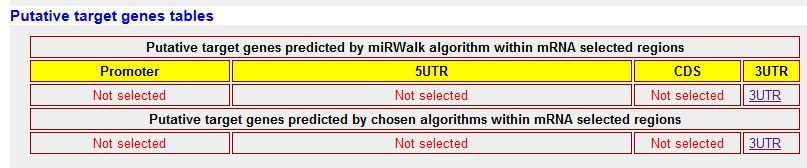 |
| This table shows only results for the selected 3'-UTR region of the genes. You have two hyperlinks below 3UTR, one corresponding to the prediction of miRWalk algorithm and other for the multiple prediction of the selected algorithms. |
After selecting the link corresponding to the chosen algorithms you will get a table with the comparative results of the target prediction by all the selected algorithms. Stronger targets should be predicted by higher number of algorithms. As a role of thumb, I normally recommend to consider as "good targets" those which are predicted by more than 2/3 of the used algorithms.
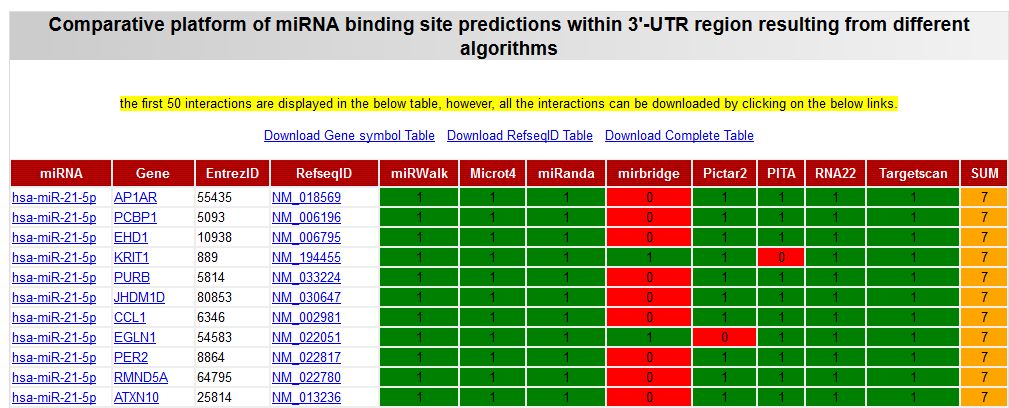 |
| Positive predictions are indicated by "1" and negative by "0". Sum of total positive predictions are in the last right column of the table. These results can be downloaded and imported into Excel by using the link "Download complete table". |
If we select to show the results obtained only by miRWalk algorithm, the corresponding table will show more detailed results, including the seed lenght and positions in the 3'-UTR. However, remember to first filter your target list using the multiple prediction.
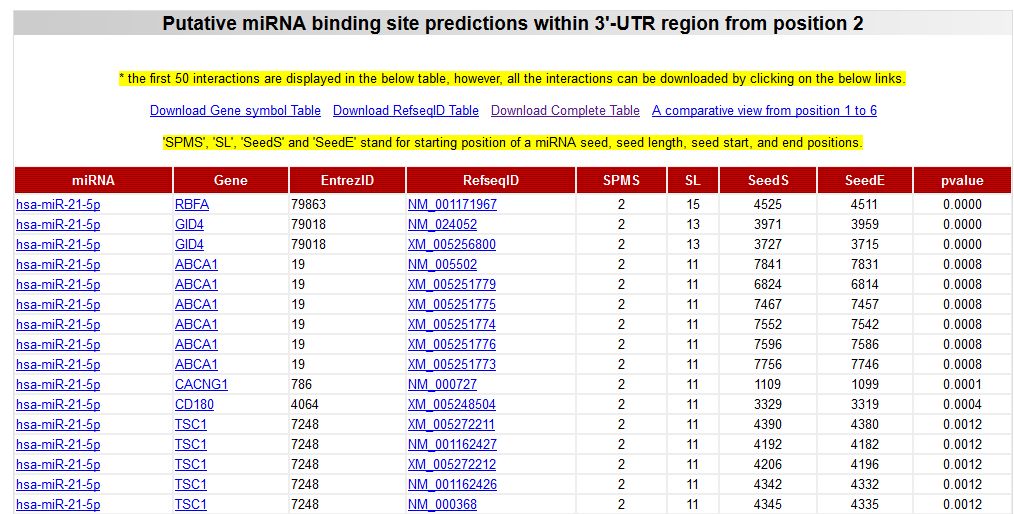 |
| The results of the miRWalk algorithm include much more information regarding the lenght of the pairing seed segment, the relative position of the predicted target in the 3'-UTR, and also the statistical significance of the prediction (p-value). Also you can see that some genes appeared many times, indicating multiple targets for hsa-miR-21-5p in their 3'-UTR. |
5.- Validated targets for hsa-miR-21-5p
For validated target retrieval we will take advantage of miRTarbase. Open the miRTarbase link and introduce "hsa-mir-21-5p" in the search window. The search will give you the following output:
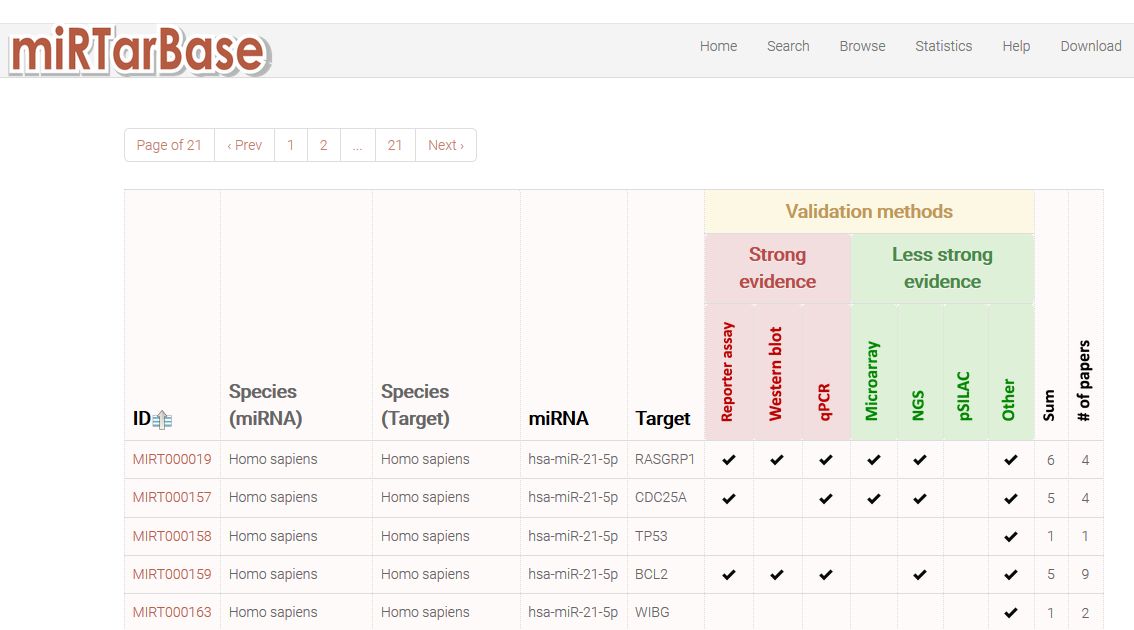 |
| These tabular results show the list of validated targets for hsa-miR21-5p, indicating the validation methods and the strenght of them. Additional data is accessible by clicking the ID of the target. In additional pages you will find information about experimental evidences, expression correlation between miRNA and its target in different cells and tissues, and also regulatory networks constructed from the validated target data. It is a very complete and cool database with lots of information. |